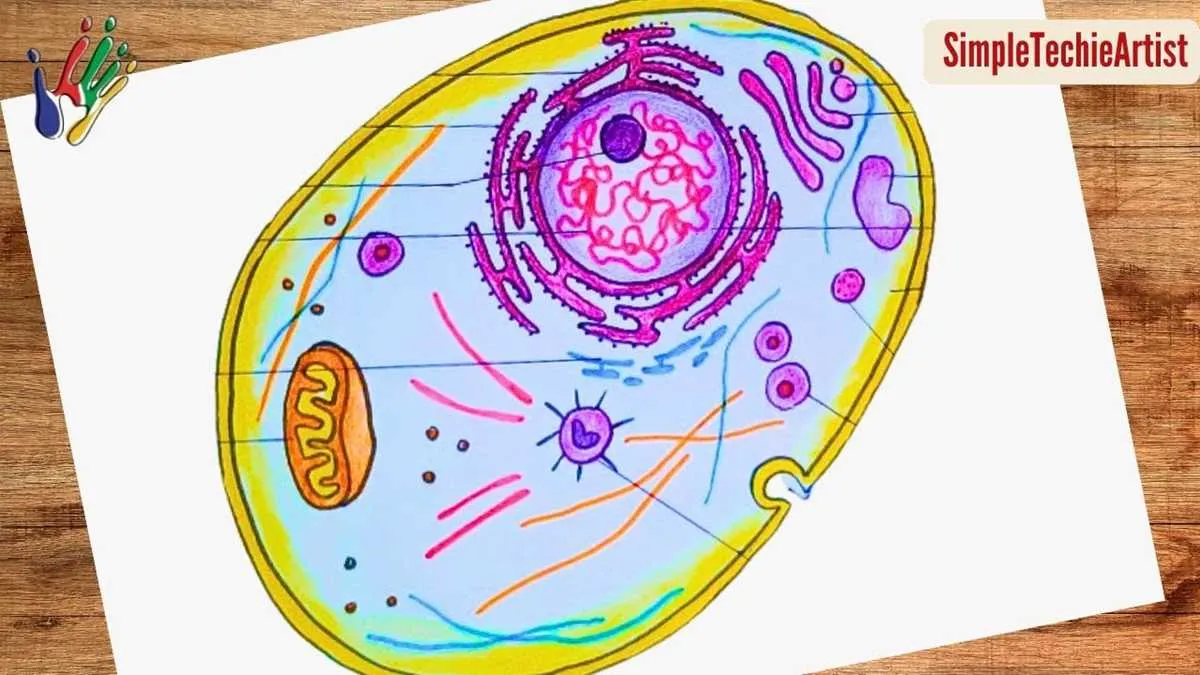
Focus on identifying key components such as the nucleus, which serves as the control center containing genetic material. Recognize the mitochondria as essential energy producers, often referred to as the powerhouse of the unit. Pay close attention to the endoplasmic reticulum, distinguished into rough and smooth types, responsible for protein synthesis and lipid metabolism respectively.
Membrane-bound structures like the Golgi apparatus play a crucial role in modifying and packaging molecules for transport. The cytoplasm fills the interior space, providing a medium for biochemical reactions and housing various organelles. Observe the lysosomes which act as waste disposal systems, digesting unwanted materials to maintain internal health.
When analyzing an unlabeled schematic, begin by correlating the shapes and locations of each component to their functions. Understanding these elements within the framework of a single living unit facilitates deeper insight into biological processes and cellular interactions at a microscopic level.
Unmarked Illustration of a Eukaryotic Cytoplasm
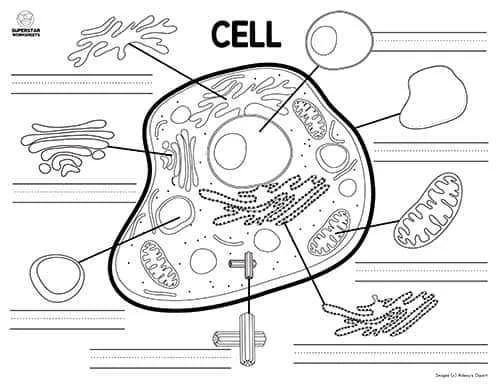
Focus on identifying key organelles by their distinct shapes and relative positions. The nucleus appears as a large, spherical structure often near the center, containing a darker nucleolus. Surrounding it, the cytoplasmic matrix houses numerous mitochondria, recognized by their double membranes and folded inner surfaces.
Look for the endoplasmic reticulum, distinguishable in two forms: rough, dotted with ribosomes, and smooth, lacking these granules. The Golgi apparatus presents as a series of stacked, flattened sacs close to the nucleus. Lysosomes and peroxisomes are smaller, spherical vesicles scattered throughout the cytoplasm.
Membrane-bound vacuoles may be present, though smaller and less prominent than in plant equivalents. The plasma membrane outlines the entire structure, appearing as a thin boundary that controls molecular exchange. Identifying cytoskeletal elements can be challenging without labels, but microtubules and filaments support the internal framework.
For effective study, compare the illustration with labeled references to train recognition by morphology and spatial arrangement rather than relying on annotations. This method enhances understanding of intracellular organization and functional relationships.
How to Identify Key Organelles Without Labels
Focus on distinct shapes and textures to recognize major internal structures. The nucleus appears as a large, spherical body often near the center, characterized by a dense double membrane and sometimes visible nucleolus inside.
Look for the mitochondria by their oval shape and the presence of inner folds called cristae, which differentiate them from other small organelles. The endoplasmic reticulum forms a network of tubular membranes; rough regions have dots representing ribosomes, while smooth parts lack them and appear more tubular.
Lysosomes are small, spherical vesicles with a uniform texture, often scattered throughout the cytoplasm. The Golgi apparatus appears as a series of flattened, stacked sacs located near the nucleus, distinct by their curved, layered arrangement.
Spot the cytoskeleton by identifying thin filaments or tubules that provide structural support; these often look like fine lines crisscrossing the interior. Vesicles are small, round compartments that transport materials and appear as tiny bubbles within the fluid matrix.
Techniques for Annotating Unlabelled Cell Diagrams
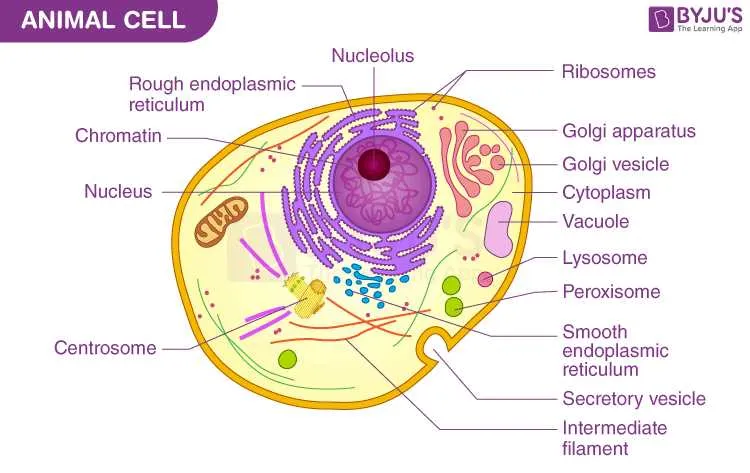
Begin by identifying key organelles through shape and location patterns recognized in typical eukaryotic microscopic visuals. Use reference charts to match structures such as the nucleus, mitochondria, endoplasmic reticulum, and Golgi apparatus.
Apply color-coding to differentiate components: assign distinct hues for the cytoplasm, membrane boundaries, and internal compartments to enhance clarity. This approach aids memory retention and visual separation.
Incorporate numbering systems alongside brief labels, linking each figure to a detailed legend. Maintain consistent numbering order, typically starting from the nucleus and moving outward, to ensure systematic annotation.
Utilize arrows and pointers to accurately connect annotations with corresponding regions, avoiding ambiguity. Precision in placement is critical for effective identification and study.
Cross-reference with microscopic images or 3D models for validation. This practice minimizes errors and reinforces accuracy in marking less obvious structures.
Common Mistakes When Interpreting Unlabelled Cell Images
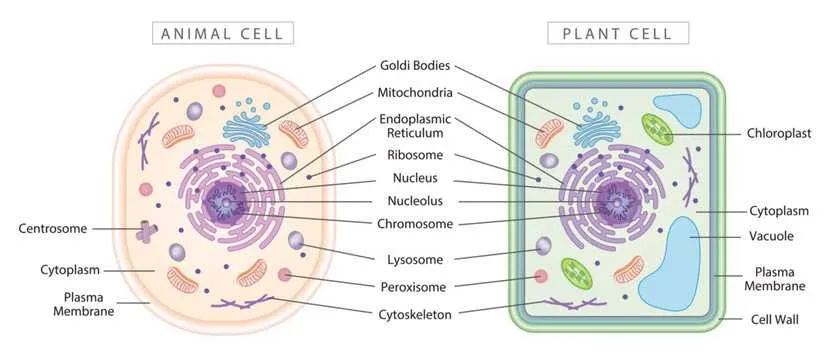
Always verify organelle shapes and relative positions before identification to avoid confusion.
- Misidentifying the nucleus: Often mistaken for vacuoles or other large structures. Remember, the nucleus is usually centrally located and contains a distinct nucleolus.
- Confusing mitochondria with lysosomes: Both can appear as small, round bodies, but mitochondria typically have a double membrane and internal folds (cristae), unlike lysosomes.
- Overlooking the cell membrane: Some images show only the cytoplasm outline, leading to misinterpretation of the cell boundary. Focus on the thin, continuous layer surrounding the cytoplasm.
- Ignoring cytoplasmic granules: Granules can be mistaken for artifacts or organelles. Look for consistent patterns and texture to distinguish them properly.
- Misreading endoplasmic reticulum: Rough ER is often confused with smooth ER; rough ER is studded with ribosomes, making it appear grainy, while smooth ER looks tubular and smooth.
Follow this checklist when analyzing unlabeled cellular images:
- Locate the largest, round structure to confirm it as the nucleus.
- Identify membrane-bound bodies and distinguish mitochondria by their internal structure.
- Trace the outer edge carefully to establish the plasma membrane.
- Note the texture differences to separate rough and smooth networks within the cytoplasm.
- Compare granule distribution patterns to known organelle maps.